Nota
Haz clic en aquí para descargar el código de ejemplo completo o para ejecutar este ejemplo en tu navegador a través de Binder
Ejercicio de validación cruzada sobre el conjunto de datos de la diabetes¶
Un ejercicio tutorial que utiliza la validación cruzada con modelos lineales.
Este ejercicio se utiliza en la parte Estimadores con validación cruzada de la sección Selección de modelos: elección de estimadores y sus parámetros del Un tutorial sobre aprendizaje estadístico para el procesamiento de datos científicos.
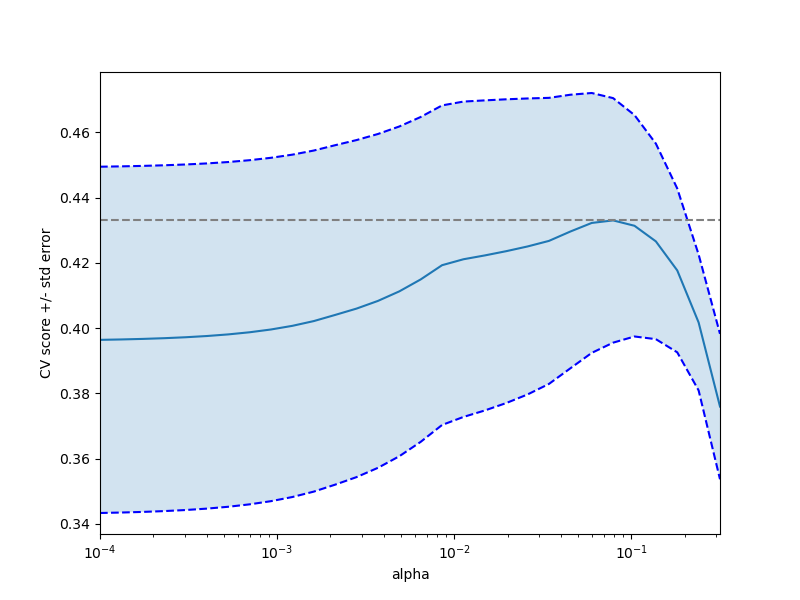
Out:
Answer to the bonus question: how much can you trust the selection of alpha?
Alpha parameters maximising the generalization score on different
subsets of the data:
[fold 0] alpha: 0.05968, score: 0.54209
[fold 1] alpha: 0.04520, score: 0.15523
[fold 2] alpha: 0.07880, score: 0.45193
Answer: Not very much since we obtained different alphas for different
subsets of the data and moreover, the scores for these alphas differ
quite substantially.
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.linear_model import LassoCV
from sklearn.linear_model import Lasso
from sklearn.model_selection import KFold
from sklearn.model_selection import GridSearchCV
X, y = datasets.load_diabetes(return_X_y=True)
X = X[:150]
y = y[:150]
lasso = Lasso(random_state=0, max_iter=10000)
alphas = np.logspace(-4, -0.5, 30)
tuned_parameters = [{'alpha': alphas}]
n_folds = 5
clf = GridSearchCV(lasso, tuned_parameters, cv=n_folds, refit=False)
clf.fit(X, y)
scores = clf.cv_results_['mean_test_score']
scores_std = clf.cv_results_['std_test_score']
plt.figure().set_size_inches(8, 6)
plt.semilogx(alphas, scores)
# plot error lines showing +/- std. errors of the scores
std_error = scores_std / np.sqrt(n_folds)
plt.semilogx(alphas, scores + std_error, 'b--')
plt.semilogx(alphas, scores - std_error, 'b--')
# alpha=0.2 controls the translucency of the fill color
plt.fill_between(alphas, scores + std_error, scores - std_error, alpha=0.2)
plt.ylabel('CV score +/- std error')
plt.xlabel('alpha')
plt.axhline(np.max(scores), linestyle='--', color='.5')
plt.xlim([alphas[0], alphas[-1]])
# #############################################################################
# Bonus: how much can you trust the selection of alpha?
# To answer this question we use the LassoCV object that sets its alpha
# parameter automatically from the data by internal cross-validation (i.e. it
# performs cross-validation on the training data it receives).
# We use external cross-validation to see how much the automatically obtained
# alphas differ across different cross-validation folds.
lasso_cv = LassoCV(alphas=alphas, random_state=0, max_iter=10000)
k_fold = KFold(3)
print("Answer to the bonus question:",
"how much can you trust the selection of alpha?")
print()
print("Alpha parameters maximising the generalization score on different")
print("subsets of the data:")
for k, (train, test) in enumerate(k_fold.split(X, y)):
lasso_cv.fit(X[train], y[train])
print("[fold {0}] alpha: {1:.5f}, score: {2:.5f}".
format(k, lasso_cv.alpha_, lasso_cv.score(X[test], y[test])))
print()
print("Answer: Not very much since we obtained different alphas for different")
print("subsets of the data and moreover, the scores for these alphas differ")
print("quite substantially.")
plt.show()
Tiempo total de ejecución del script: (0 minutos 0.677 segundos)
