Nota
Haz clic en aquí para descargar el código de ejemplo completo o para ejecutar este ejemplo en tu navegador a través de Binder
Ejercicio SVM¶
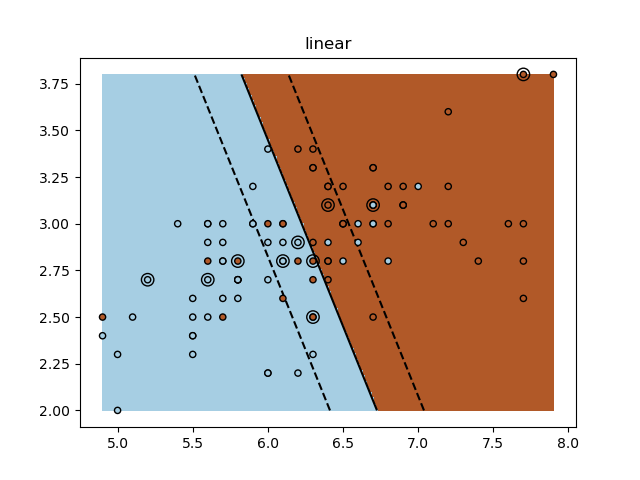
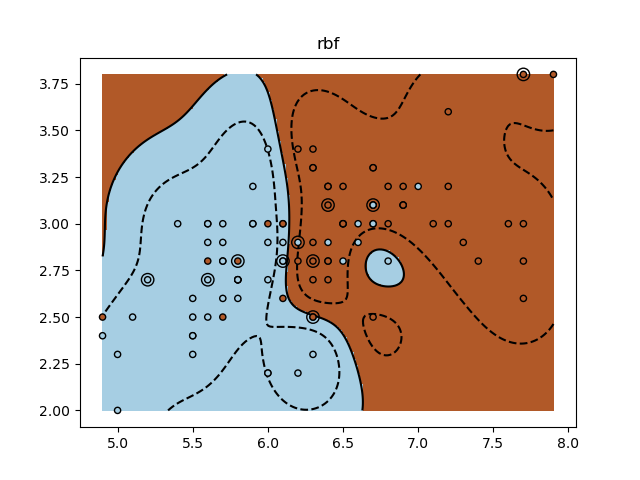
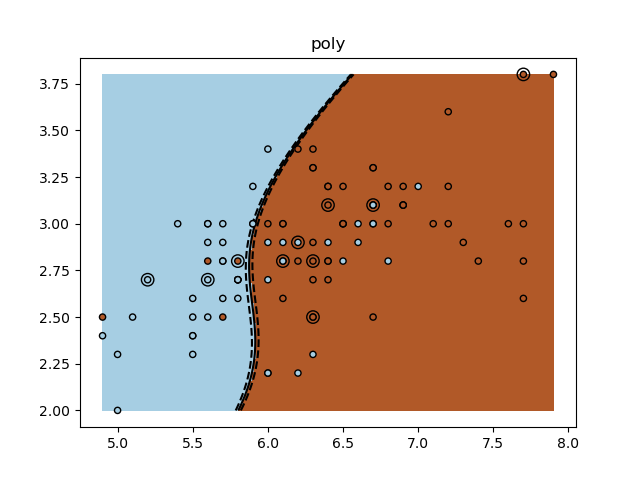
Un ejercicio tutorial para utilizar diferentes núcleos de SVM.
Este ejercicio se utiliza en la parte Uso de los núcleos (kernels) de la sección Aprendizaje supervisado: predicción de una variable de salida a partir de observaciones de alta dimensión del Un tutorial sobre aprendizaje estadístico para el procesamiento de datos científicos.
Out:
/home/mapologo/Descargas/scikit-learn-0.24.X/examples/exercises/plot_iris_exercise.py:62: MatplotlibDeprecationWarning: shading='flat' when X and Y have the same dimensions as C is deprecated since 3.3. Either specify the corners of the quadrilaterals with X and Y, or pass shading='auto', 'nearest' or 'gouraud', or set rcParams['pcolor.shading']. This will become an error two minor releases later.
plt.pcolormesh(XX, YY, Z > 0, cmap=plt.cm.Paired)
/home/mapologo/Descargas/scikit-learn-0.24.X/examples/exercises/plot_iris_exercise.py:62: MatplotlibDeprecationWarning: shading='flat' when X and Y have the same dimensions as C is deprecated since 3.3. Either specify the corners of the quadrilaterals with X and Y, or pass shading='auto', 'nearest' or 'gouraud', or set rcParams['pcolor.shading']. This will become an error two minor releases later.
plt.pcolormesh(XX, YY, Z > 0, cmap=plt.cm.Paired)
/home/mapologo/Descargas/scikit-learn-0.24.X/examples/exercises/plot_iris_exercise.py:62: MatplotlibDeprecationWarning: shading='flat' when X and Y have the same dimensions as C is deprecated since 3.3. Either specify the corners of the quadrilaterals with X and Y, or pass shading='auto', 'nearest' or 'gouraud', or set rcParams['pcolor.shading']. This will become an error two minor releases later.
plt.pcolormesh(XX, YY, Z > 0, cmap=plt.cm.Paired)
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets, svm
iris = datasets.load_iris()
X = iris.data
y = iris.target
X = X[y != 0, :2]
y = y[y != 0]
n_sample = len(X)
np.random.seed(0)
order = np.random.permutation(n_sample)
X = X[order]
y = y[order].astype(float)
X_train = X[:int(.9 * n_sample)]
y_train = y[:int(.9 * n_sample)]
X_test = X[int(.9 * n_sample):]
y_test = y[int(.9 * n_sample):]
# fit the model
for kernel in ('linear', 'rbf', 'poly'):
clf = svm.SVC(kernel=kernel, gamma=10)
clf.fit(X_train, y_train)
plt.figure()
plt.clf()
plt.scatter(X[:, 0], X[:, 1], c=y, zorder=10, cmap=plt.cm.Paired,
edgecolor='k', s=20)
# Circle out the test data
plt.scatter(X_test[:, 0], X_test[:, 1], s=80, facecolors='none',
zorder=10, edgecolor='k')
plt.axis('tight')
x_min = X[:, 0].min()
x_max = X[:, 0].max()
y_min = X[:, 1].min()
y_max = X[:, 1].max()
XX, YY = np.mgrid[x_min:x_max:200j, y_min:y_max:200j]
Z = clf.decision_function(np.c_[XX.ravel(), YY.ravel()])
# Put the result into a color plot
Z = Z.reshape(XX.shape)
plt.pcolormesh(XX, YY, Z > 0, cmap=plt.cm.Paired)
plt.contour(XX, YY, Z, colors=['k', 'k', 'k'],
linestyles=['--', '-', '--'], levels=[-.5, 0, .5])
plt.title(kernel)
plt.show()
Tiempo total de ejecución del script: (0 minutos 6.606 segundos)