Nota
Haga clic en aquí para descargar el código completo del ejemplo o para ejecutar este ejemplo en su navegador a través de Binder
Extracción de temas con Factorización Matricial No Negativa y Asignación de Dirichlet Latente¶
Este es un ejemplo de aplicación de NMF y LatentDirichletAllocation sobre un corpus de documentos y extraer modelos aditivos de la estructura de temas del corpus. El resultado es un diagrama de temas, cada uno representado como un diagrama de barras utilizando las palabras más importantes en función de las ponderaciones.
La factorización de matrices no negativas se aplica con dos funciones objetivo diferentes: la norma de Frobenius y la divergencia de Kullback-Leibler generalizada. Esta última es equivalente a la indexación semántica latente probabilística.
Los parámetros por defecto (n_samples / n_features / n_components) deberían hacer que el ejemplo se pueda ejecutar en un par de decenas de segundos. Puedes intentar aumentar las dimensiones del problema, pero ten en cuenta que la complejidad temporal es polinómica en NMF. En LDA, la complejidad de tiempo es proporcional a (n_samples * iterations).
Out:
Loading dataset...
done in 1.443s.
Extracting tf-idf features for NMF...
done in 0.321s.
Extracting tf features for LDA...
done in 0.463s.
Fitting the NMF model (Frobenius norm) with tf-idf features, n_samples=2000 and n_features=1000...
/home/mapologo/miniconda3/envs/sklearn/lib/python3.9/site-packages/scikit_learn-0.24.1-py3.9-linux-x86_64.egg/sklearn/decomposition/_nmf.py:312: FutureWarning: The 'init' value, when 'init=None' and n_components is less than n_samples and n_features, will be changed from 'nndsvd' to 'nndsvda' in 1.1 (renaming of 0.26).
warnings.warn(("The 'init' value, when 'init=None' and "
done in 0.300s.
Fitting the NMF model (generalized Kullback-Leibler divergence) with tf-idf features, n_samples=2000 and n_features=1000...
/home/mapologo/miniconda3/envs/sklearn/lib/python3.9/site-packages/scikit_learn-0.24.1-py3.9-linux-x86_64.egg/sklearn/decomposition/_nmf.py:312: FutureWarning: The 'init' value, when 'init=None' and n_components is less than n_samples and n_features, will be changed from 'nndsvd' to 'nndsvda' in 1.1 (renaming of 0.26).
warnings.warn(("The 'init' value, when 'init=None' and "
done in 1.382s.
Fitting LDA models with tf features, n_samples=2000 and n_features=1000...
done in 3.912s.
# Author: Olivier Grisel <olivier.grisel@ensta.org>
# Lars Buitinck
# Chyi-Kwei Yau <chyikwei.yau@gmail.com>
# License: BSD 3 clause
from time import time
import matplotlib.pyplot as plt
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
from sklearn.decomposition import NMF, LatentDirichletAllocation
from sklearn.datasets import fetch_20newsgroups
n_samples = 2000
n_features = 1000
n_components = 10
n_top_words = 20
def plot_top_words(model, feature_names, n_top_words, title):
fig, axes = plt.subplots(2, 5, figsize=(30, 15), sharex=True)
axes = axes.flatten()
for topic_idx, topic in enumerate(model.components_):
top_features_ind = topic.argsort()[:-n_top_words - 1:-1]
top_features = [feature_names[i] for i in top_features_ind]
weights = topic[top_features_ind]
ax = axes[topic_idx]
ax.barh(top_features, weights, height=0.7)
ax.set_title(f'Topic {topic_idx +1}',
fontdict={'fontsize': 30})
ax.invert_yaxis()
ax.tick_params(axis='both', which='major', labelsize=20)
for i in 'top right left'.split():
ax.spines[i].set_visible(False)
fig.suptitle(title, fontsize=40)
plt.subplots_adjust(top=0.90, bottom=0.05, wspace=0.90, hspace=0.3)
plt.show()
# Load the 20 newsgroups dataset and vectorize it. We use a few heuristics
# to filter out useless terms early on: the posts are stripped of headers,
# footers and quoted replies, and common English words, words occurring in
# only one document or in at least 95% of the documents are removed.
print("Loading dataset...")
t0 = time()
data, _ = fetch_20newsgroups(shuffle=True, random_state=1,
remove=('headers', 'footers', 'quotes'),
return_X_y=True)
data_samples = data[:n_samples]
print("done in %0.3fs." % (time() - t0))
# Use tf-idf features for NMF.
print("Extracting tf-idf features for NMF...")
tfidf_vectorizer = TfidfVectorizer(max_df=0.95, min_df=2,
max_features=n_features,
stop_words='english')
t0 = time()
tfidf = tfidf_vectorizer.fit_transform(data_samples)
print("done in %0.3fs." % (time() - t0))
# Use tf (raw term count) features for LDA.
print("Extracting tf features for LDA...")
tf_vectorizer = CountVectorizer(max_df=0.95, min_df=2,
max_features=n_features,
stop_words='english')
t0 = time()
tf = tf_vectorizer.fit_transform(data_samples)
print("done in %0.3fs." % (time() - t0))
print()
# Fit the NMF model
print("Fitting the NMF model (Frobenius norm) with tf-idf features, "
"n_samples=%d and n_features=%d..."
% (n_samples, n_features))
t0 = time()
nmf = NMF(n_components=n_components, random_state=1,
alpha=.1, l1_ratio=.5).fit(tfidf)
print("done in %0.3fs." % (time() - t0))
tfidf_feature_names = tfidf_vectorizer.get_feature_names()
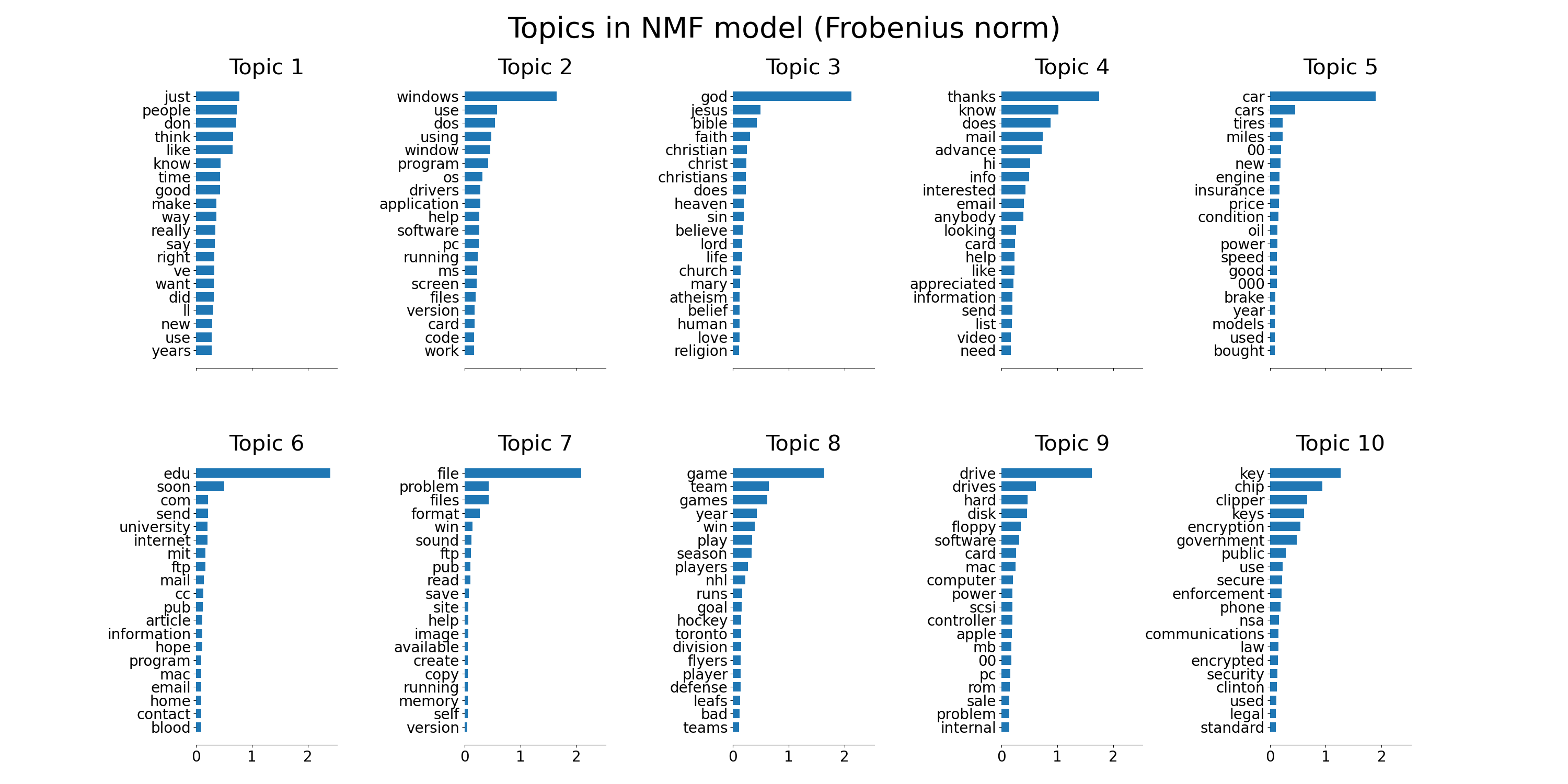
plot_top_words(nmf, tfidf_feature_names, n_top_words,
'Topics in NMF model (Frobenius norm)')
# Fit the NMF model
print('\n' * 2, "Fitting the NMF model (generalized Kullback-Leibler "
"divergence) with tf-idf features, n_samples=%d and n_features=%d..."
% (n_samples, n_features))
t0 = time()
nmf = NMF(n_components=n_components, random_state=1,
beta_loss='kullback-leibler', solver='mu', max_iter=1000, alpha=.1,
l1_ratio=.5).fit(tfidf)
print("done in %0.3fs." % (time() - t0))
tfidf_feature_names = tfidf_vectorizer.get_feature_names()
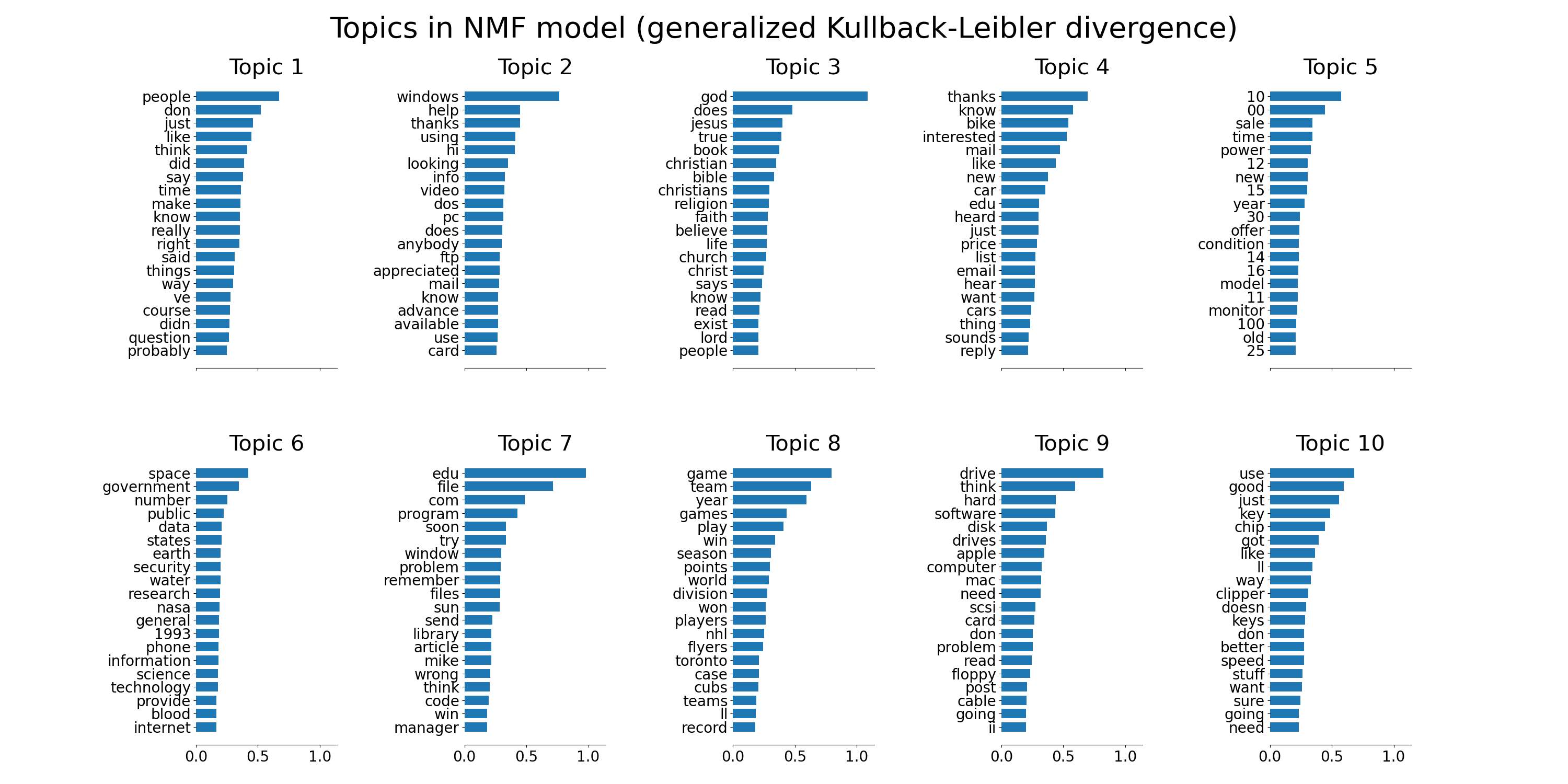
plot_top_words(nmf, tfidf_feature_names, n_top_words,
'Topics in NMF model (generalized Kullback-Leibler divergence)')
print('\n' * 2, "Fitting LDA models with tf features, "
"n_samples=%d and n_features=%d..."
% (n_samples, n_features))
lda = LatentDirichletAllocation(n_components=n_components, max_iter=5,
learning_method='online',
learning_offset=50.,
random_state=0)
t0 = time()
lda.fit(tf)
print("done in %0.3fs." % (time() - t0))
tf_feature_names = tf_vectorizer.get_feature_names()
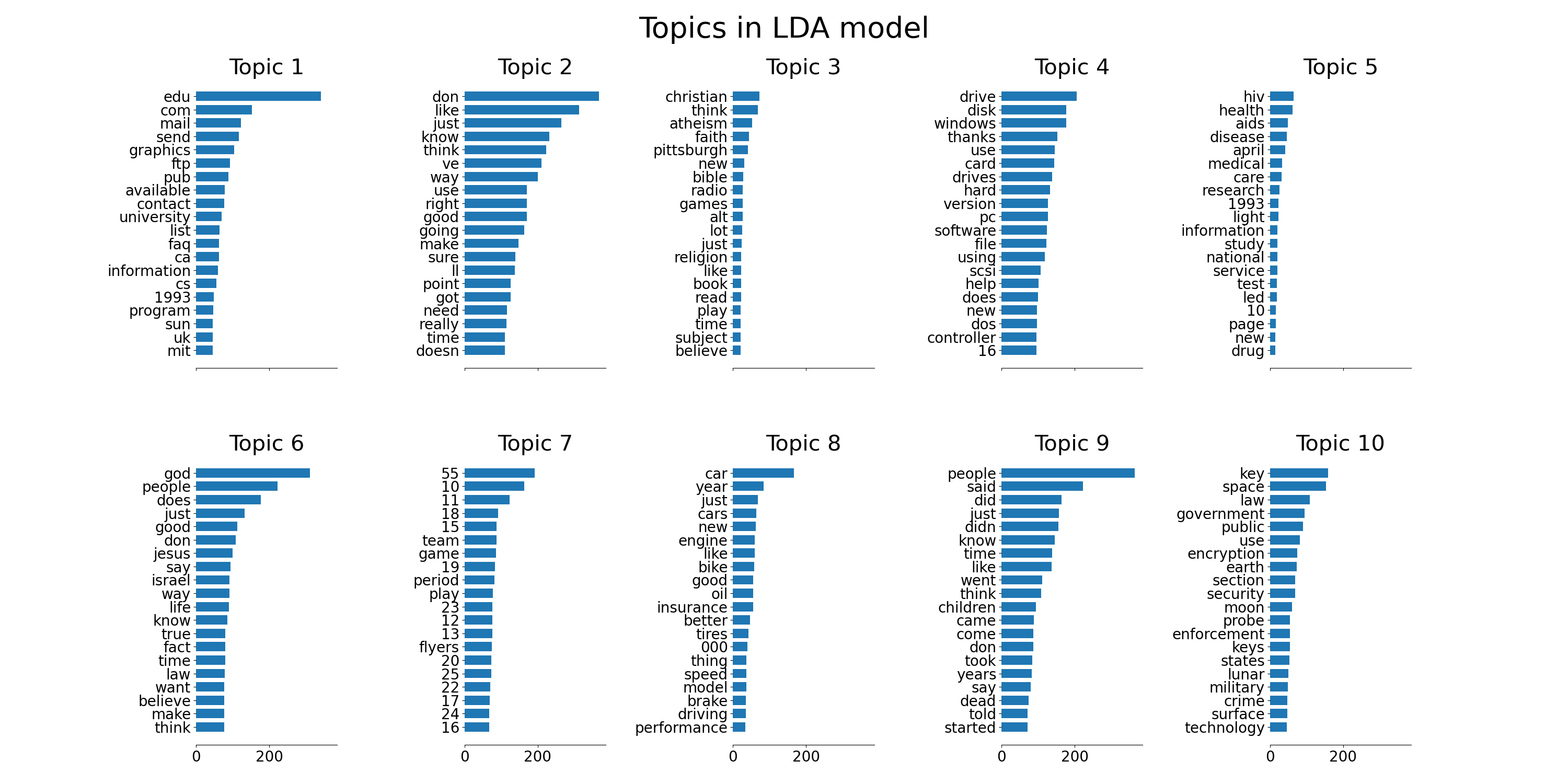
plot_top_words(lda, tf_feature_names, n_top_words, 'Topics in LDA model')
Tiempo total de ejecución del script: (0 minutos 13.176 segundos)