Nota
Haz clic aquí para descargar el código completo del ejemplo o para ejecutar este ejemplo en tu navegador a través de Binder
Ajuste al azar en la evaluación del rendimiento del agrupamiento¶
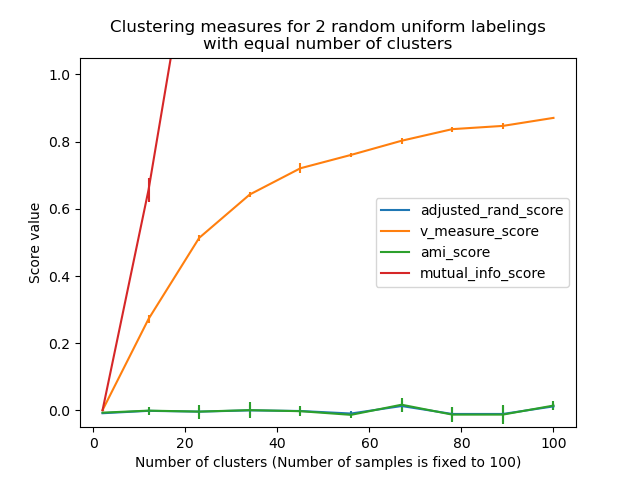
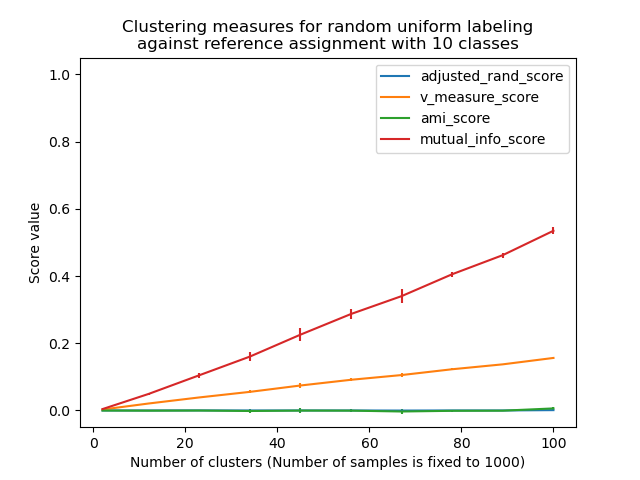
Los siguientes gráficos demuestran el impacto del número de conglomerados y del número de muestras en varias métricas de evaluación del rendimiento del agrupamiento.
Las medidas no ajustadas, como la Medida V, muestran una dependencia entre el número de conglomerados y el número de muestras: la media de la Medida V del etiquetado aleatorio aumenta significativamente a medida que el número de conglomerados se acerca al número total de muestras utilizadas para calcular la medida.
Las medidas ajustadas al azar, como el ARI, muestran algunas variaciones aleatorias centradas en una puntuación media de 0.0 para cualquier número de muestras y conglomerados.
Por tanto, sólo las medidas ajustadas pueden utilizarse con seguridad como índice de consenso para evaluar la estabilidad promedio de los algoritmos de agrupamiento para un valor determinado de k en varias submuestras superpuestas del conjunto de datos.
Out:
Computing adjusted_rand_score for 10 values of n_clusters and n_samples=100
done in 0.039s
Computing v_measure_score for 10 values of n_clusters and n_samples=100
done in 0.059s
Computing ami_score for 10 values of n_clusters and n_samples=100
done in 0.356s
Computing mutual_info_score for 10 values of n_clusters and n_samples=100
done in 0.049s
Computing adjusted_rand_score for 10 values of n_clusters and n_samples=1000
done in 0.051s
Computing v_measure_score for 10 values of n_clusters and n_samples=1000
done in 0.087s
Computing ami_score for 10 values of n_clusters and n_samples=1000
done in 0.255s
Computing mutual_info_score for 10 values of n_clusters and n_samples=1000
done in 0.062s
print(__doc__)
# Author: Olivier Grisel <olivier.grisel@ensta.org>
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
from time import time
from sklearn import metrics
def uniform_labelings_scores(score_func, n_samples, n_clusters_range,
fixed_n_classes=None, n_runs=5, seed=42):
"""Compute score for 2 random uniform cluster labelings.
Both random labelings have the same number of clusters for each value
possible value in ``n_clusters_range``.
When fixed_n_classes is not None the first labeling is considered a ground
truth class assignment with fixed number of classes.
"""
random_labels = np.random.RandomState(seed).randint
scores = np.zeros((len(n_clusters_range), n_runs))
if fixed_n_classes is not None:
labels_a = random_labels(low=0, high=fixed_n_classes, size=n_samples)
for i, k in enumerate(n_clusters_range):
for j in range(n_runs):
if fixed_n_classes is None:
labels_a = random_labels(low=0, high=k, size=n_samples)
labels_b = random_labels(low=0, high=k, size=n_samples)
scores[i, j] = score_func(labels_a, labels_b)
return scores
def ami_score(U, V):
return metrics.adjusted_mutual_info_score(U, V)
score_funcs = [
metrics.adjusted_rand_score,
metrics.v_measure_score,
ami_score,
metrics.mutual_info_score,
]
# 2 independent random clusterings with equal cluster number
n_samples = 100
n_clusters_range = np.linspace(2, n_samples, 10).astype(int)
plt.figure(1)
plots = []
names = []
for score_func in score_funcs:
print("Computing %s for %d values of n_clusters and n_samples=%d"
% (score_func.__name__, len(n_clusters_range), n_samples))
t0 = time()
scores = uniform_labelings_scores(score_func, n_samples, n_clusters_range)
print("done in %0.3fs" % (time() - t0))
plots.append(plt.errorbar(
n_clusters_range, np.median(scores, axis=1), scores.std(axis=1))[0])
names.append(score_func.__name__)
plt.title("Clustering measures for 2 random uniform labelings\n"
"with equal number of clusters")
plt.xlabel('Number of clusters (Number of samples is fixed to %d)' % n_samples)
plt.ylabel('Score value')
plt.legend(plots, names)
plt.ylim(bottom=-0.05, top=1.05)
# Random labeling with varying n_clusters against ground class labels
# with fixed number of clusters
n_samples = 1000
n_clusters_range = np.linspace(2, 100, 10).astype(int)
n_classes = 10
plt.figure(2)
plots = []
names = []
for score_func in score_funcs:
print("Computing %s for %d values of n_clusters and n_samples=%d"
% (score_func.__name__, len(n_clusters_range), n_samples))
t0 = time()
scores = uniform_labelings_scores(score_func, n_samples, n_clusters_range,
fixed_n_classes=n_classes)
print("done in %0.3fs" % (time() - t0))
plots.append(plt.errorbar(
n_clusters_range, scores.mean(axis=1), scores.std(axis=1))[0])
names.append(score_func.__name__)
plt.title("Clustering measures for random uniform labeling\n"
"against reference assignment with %d classes" % n_classes)
plt.xlabel('Number of clusters (Number of samples is fixed to %d)' % n_samples)
plt.ylabel('Score value')
plt.ylim(bottom=-0.05, top=1.05)
plt.legend(plots, names)
plt.show()
Tiempo total de ejecución del script: (0 minutos 1.202 segundos)